Quickstart Guide
Get up and running with Pulseq on Philips in three simple steps.
Prerequisites
Before you begin, ensure you have:
- New to Pulseq? Check out the Pulseq Tutorials
- New to Philips? There are courses provided by Philips and Gyrotools
No MATLAB or Python?
You can still follow along by downloading the example .seq files directly, or by using an online environment:
Step 1: Get a Sequence File
You can either download a pre-made sequence file or create your own:
Start with one of our validated examples.
View ExamplesUse PyPulseq or MATLAB to design a sequence.
Pulseq Tutorials
Minimal Example
Here's a simple Gradient Echo (GRE) sequence to get you started.
import math
import numpy as np
import pypulseq as pp
def main(plot: bool = False, write_seq: bool = False, seq_filename: str = 'gre_pypulseq.seq'):
# ======
# SETUP
# ======
# Create a new sequence object
fov = 256e-3 # Define FOV and resolution
Nx = 64
Ny = 64
alpha = 10 # flip angle
slice_thickness = 3e-3 # slice
TR = 12e-3 # Repetition time
TE = 5e-3 # Echo time
rf_spoiling_inc = 117 # RF spoiling increment
system = pp.Opts(
max_grad=28,
grad_unit='mT/m',
max_slew=150,
slew_unit='T/m/s',
rf_ringdown_time=20e-6,
rf_dead_time=100e-6,
adc_dead_time=10e-6,
)
seq = pp.Sequence(system)
# ======
# CREATE EVENTS
# ======
rf, gz, _ = pp.make_sinc_pulse(
flip_angle=alpha * math.pi / 180,
duration=3e-3,
slice_thickness=slice_thickness,
apodization=0.42,
time_bw_product=4,
system=system,
return_gz=True,
delay=system.rf_dead_time,
)
# Define other gradients and ADC events
delta_k = 1 / fov
gx = pp.make_trapezoid(channel='x', flat_area=Nx * delta_k, flat_time=3.2e-3, system=system)
adc = pp.make_adc(num_samples=Nx, duration=gx.flat_time, delay=gx.rise_time, system=system)
gx_pre = pp.make_trapezoid(channel='x', area=-gx.area / 2, duration=1e-3, system=system)
gz_reph = pp.make_trapezoid(channel='z', area=-gz.area / 2, duration=1e-3, system=system)
phase_areas = (np.arange(Ny) - Ny / 2) * delta_k
# gradient spoiling
gx_spoil = pp.make_trapezoid(channel='x', area=2 * Nx * delta_k, system=system)
gz_spoil = pp.make_trapezoid(channel='z', area=4 / slice_thickness, system=system)
# Calculate timing
delay_TE = (
math.ceil(
(
TE
- (pp.calc_duration(gz, rf) - pp.calc_rf_center(rf)[0] - rf.delay)
- pp.calc_duration(gx_pre)
- pp.calc_duration(gx) / 2
- pp.eps
)
/ seq.grad_raster_time
)
* seq.grad_raster_time
)
delay_TR = (
np.ceil(
(TR - pp.calc_duration(gz, rf) - pp.calc_duration(gx_pre) - pp.calc_duration(gx) - delay_TE)
/ seq.grad_raster_time
)
* seq.grad_raster_time
)
assert np.all(delay_TE >= 0)
assert np.all(delay_TR >= pp.calc_duration(gx_spoil, gz_spoil))
rf_phase = 0
rf_inc = 0
# ======
# CONSTRUCT SEQUENCE
# ======
# Loop over phase encodes and define sequence blocks
for i in range(Ny):
rf.phase_offset = rf_phase / 180 * np.pi
adc.phase_offset = rf_phase / 180 * np.pi
rf_inc = divmod(rf_inc + rf_spoiling_inc, 360.0)[1]
rf_phase = divmod(rf_phase + rf_inc, 360.0)[1]
seq.add_block(rf, gz)
gy_pre = pp.make_trapezoid(
channel='y',
area=phase_areas[i],
duration=pp.calc_duration(gx_pre),
system=system,
)
seq.add_block(gx_pre, gy_pre, gz_reph)
seq.add_block(pp.make_delay(delay_TE))
seq.add_block(gx, adc)
gy_pre.amplitude = -gy_pre.amplitude
seq.add_block(pp.make_delay(delay_TR), gx_spoil, gy_pre, gz_spoil)
# Check whether the timing of the sequence is correct
ok, error_report = seq.check_timing()
if ok:
print('Timing check passed successfully')
else:
print('Timing check failed. Error listing follows:')
[print(e) for e in error_report]
# ======
# VISUALIZATION
# ======
if plot:
seq.plot()
seq.calculate_kspace()
# Very optional slow step, but useful for testing during development e.g. for the real TE, TR or for staying within
# slew-rate limits
rep = seq.test_report()
print(rep)
# =========
# WRITE .SEQ
# =========
if write_seq:
# Prepare the sequence output for the scanner
seq.set_definition(key='FOV', value=[fov, fov, slice_thickness])
seq.set_definition(key='Name', value='gre')
seq.write(seq_filename)
return seq
if __name__ == '__main__':
main(plot=False, write_seq=True)
% set system limits
sys = mr.opts('MaxGrad', 22, 'GradUnit', 'mT/m', ...
'MaxSlew', 120, 'SlewUnit', 'T/m/s', ...
'rfRingdownTime', 20e-6, 'rfDeadTime', 100e-6, 'adcDeadTime', 10e-6);
seq=mr.Sequence(sys); % Create a new sequence object
fov=256e-3; Nx=128; Ny=Nx; % Define FOV and resolution
alpha=10; % flip angle
sliceThickness=3e-3; % slice
TR=12e-3; % repetition time TR
TE=5e-3; % echo time TE
%TE=[7.38 9.84]*1e-3; % alternatively give a vector here to have multiple TEs (e.g. for field mapping)
% more in-depth parameters
rfSpoilingInc=117; % RF spoiling increment
roDuration=3.2e-3; % ADC duration
% Create fat-sat pulse
% (in Siemens interpreter from January 2019 duration is limited to 8.192 ms, and although product EPI uses 10.24 ms, 8 ms seems to be sufficient)
% B0=2.89; % 1.5 2.89 3.0
% sat_ppm=-3.45;
% sat_freq=sat_ppm*1e-6*B0*lims.gamma;
% rf_fs = mr.makeGaussPulse(110*pi/180,'system',lims,'Duration',8e-3,...
% 'bandwidth',abs(sat_freq),'freqOffset',sat_freq);
% gz_fs = mr.makeTrapezoid('z',sys,'delay',mr.calcDuration(rf_fs),'Area',1/1e-4); % spoil up to 0.1mm
% Create alpha-degree slice selection pulse and gradient
[rf, gz] = mr.makeSincPulse(alpha*pi/180,sys,'Duration',3e-3,...
'SliceThickness',sliceThickness,'apodization',0.42,'timeBwProduct',4,...
'use','excitation');
% Define other gradients and ADC events
deltak=1/fov;
gx = mr.makeTrapezoid('x',sys,'FlatArea',Nx*deltak,'FlatTime',roDuration);
adc = mr.makeAdc(Nx,sys,'Duration',gx.flatTime,'Delay',gx.riseTime);
gxPre = mr.makeTrapezoid('x',sys,'Area',-gx.area/2,'Duration',1e-3);
gzReph = mr.makeTrapezoid('z',sys,'Area',-gz.area/2,'Duration',1e-3);
phaseAreas = ((0:Ny-1)-Ny/2)*deltak;
gyPre = mr.makeTrapezoid('y',sys,'Area',max(abs(phaseAreas)),'Duration',mr.calcDuration(gxPre));
peScales=phaseAreas/gyPre.area;
% gradient spoiling
gxSpoil=mr.makeTrapezoid('x',sys,'Area',2*Nx*deltak);
gzSpoil=mr.makeTrapezoid('z',sys,'Area',4/sliceThickness);
% Calculate timing
delayTE=ceil((TE - mr.calcDuration(gxPre) - gz.fallTime - gz.flatTime/2 ...
- mr.calcDuration(gx)/2)/seq.gradRasterTime)*seq.gradRasterTime;
delayTR=ceil((TR - mr.calcDuration(gz) - mr.calcDuration(gxPre) ...
- mr.calcDuration(gx) - delayTE)/seq.gradRasterTime)*seq.gradRasterTime;
assert(all(delayTE>=0));
assert(all(delayTR>=mr.calcDuration(gxSpoil,gzSpoil)));
rf_phase=0;
rf_inc=0;
% Loop over phase encodes and define sequence blocks
for i=1:Ny
for c=1:length(TE)
%seq.addBlock(rf_fs,gz_fs); % fat-sat
rf.phaseOffset=rf_phase/180*pi;
adc.phaseOffset=rf_phase/180*pi;
rf_inc=mod(rf_inc+rfSpoilingInc, 360.0);
rf_phase=mod(rf_phase+rf_inc, 360.0);
%
seq.addBlock(rf,gz);
seq.addBlock(gxPre,mr.scaleGrad(gyPre,peScales(i)),gzReph);
seq.addBlock(mr.makeDelay(delayTE(c)));
seq.addBlock(gx,adc);
%gyPre.amplitude=-gyPre.amplitude;
seq.addBlock(mr.makeDelay(delayTR(c)),gxSpoil,mr.scaleGrad(gyPre,-peScales(i)),gzSpoil)
end
end
%% check whether the timing of the sequence is correct
[ok, error_report]=seq.checkTiming;
if (ok)
fprintf('Timing check passed successfully\n');
else
fprintf('Timing check failed! Error listing follows:\n');
fprintf([error_report{:}]);
fprintf('\n');
end
%% prepare sequence export
seq.setDefinition('FOV', [fov fov sliceThickness]);
seq.setDefinition('Name', 'gre');
seq.write('gre.seq') % Write to pulseq file
Step 2: Install the Interpreter
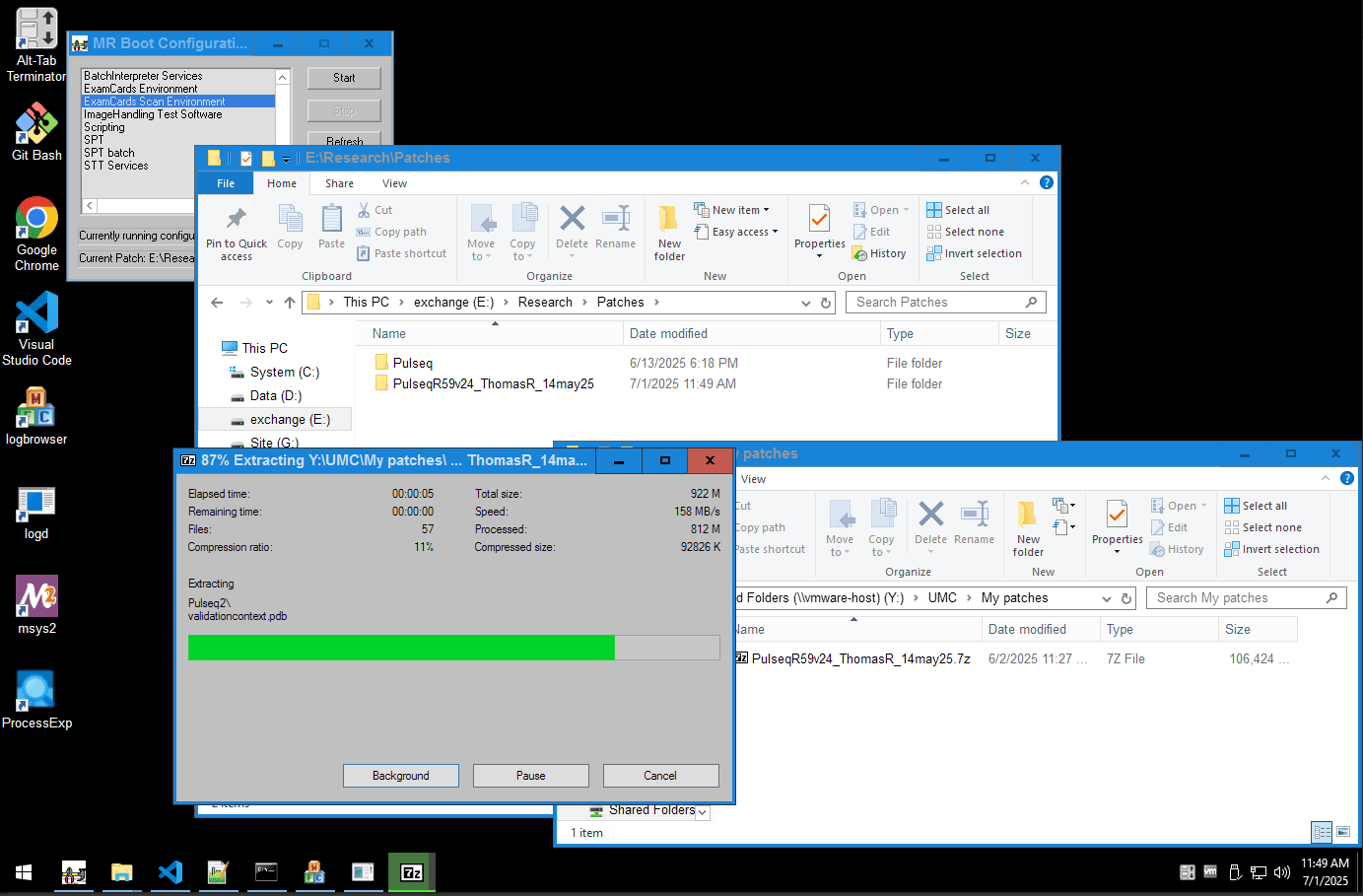
The interpreter is a Philips patch that you need to build and install.
The details on this process are available in the private GitHub repository
Research patches
The Pulseq interpreter is a patch, like any other. Patches can be wonderful, and might bring great features to your scanner.
However this patch, like others, is not made by Philips. You need to build the patch yourself, and you are responsible for its validation and usage.
Build and Install overview
- Get the code by adding the private GitHub repository as a new remote
- Checkout the interpreter branch if it is available for your scanner's relase
- If needed merge the code, into your own custom patch or branch
- Compile the code into a patch
- Test the patch and your
.seqfiles in the simulator - All okay? Copy over and enjoy the interpreter on your scanner!
More detailed instructions are available in the repository's README.
Follow all steps, including testing!
The above steps are a high-level overview. We strongly recommend following all steps in the provided README file.
If you're an experienced researcher, you might be tempted to skip some steps. Don't!
Every step is not only designed to take all safety precautions into account - they are also advised to simply provide you with the best experience.
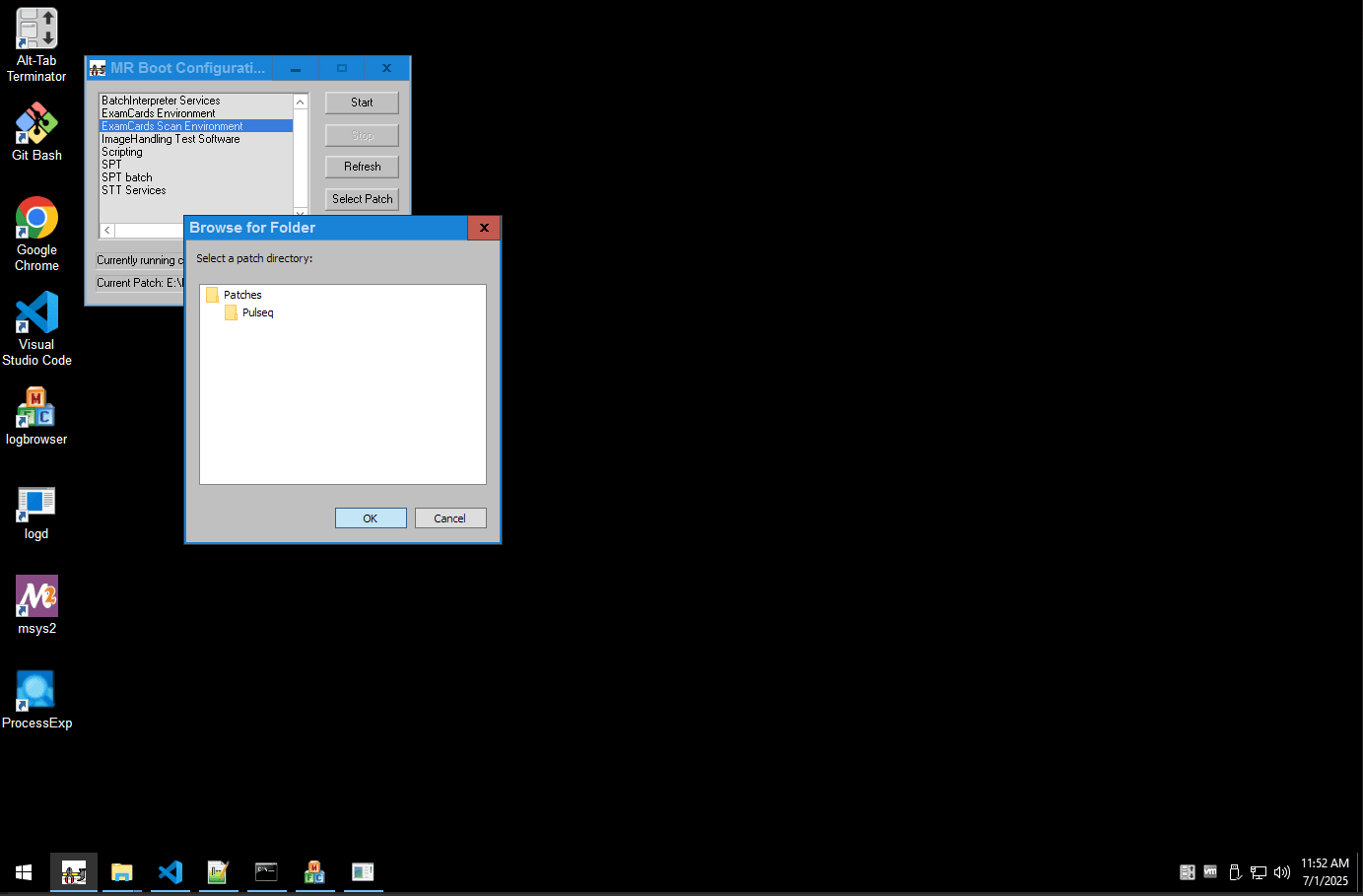
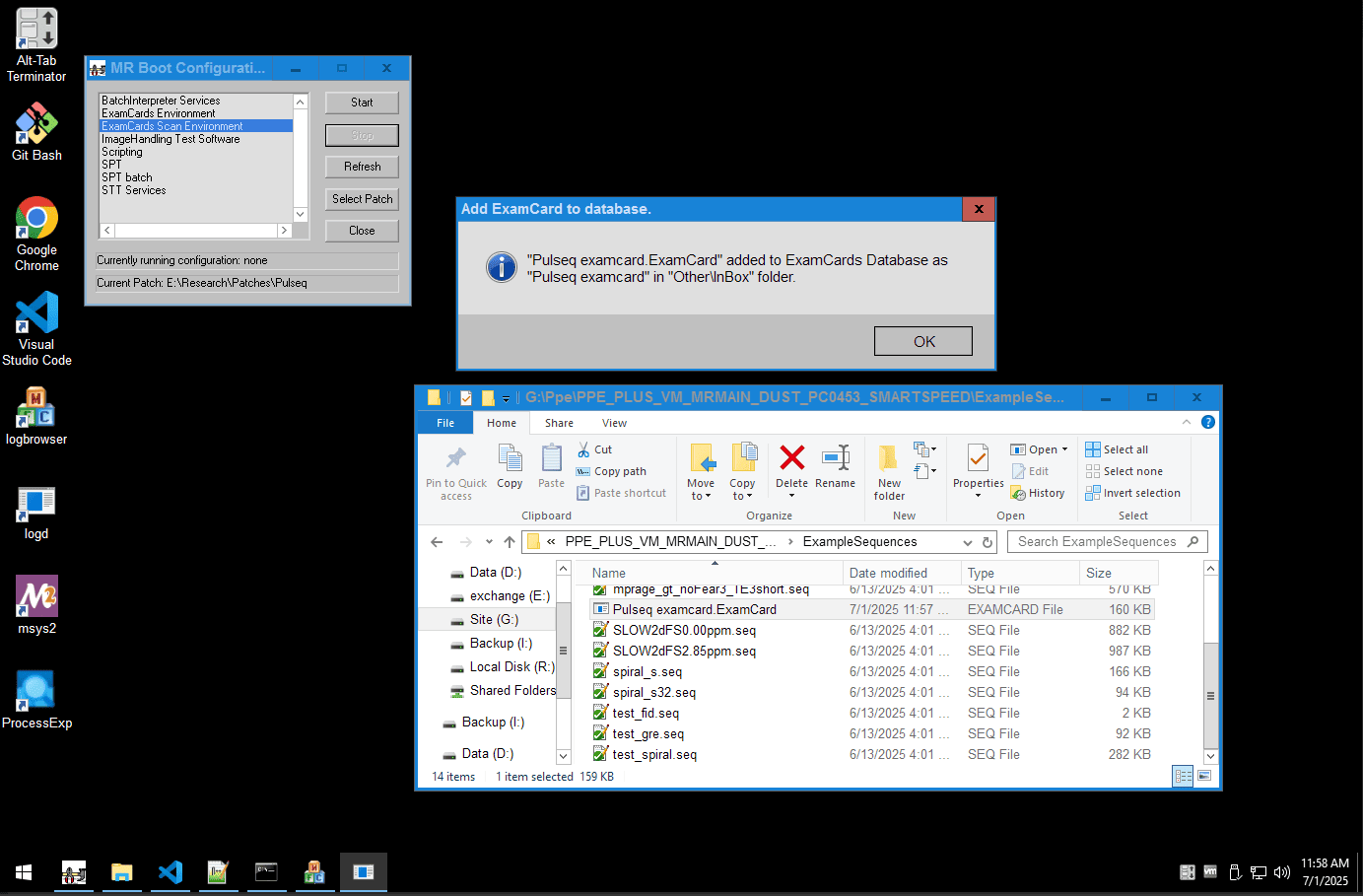
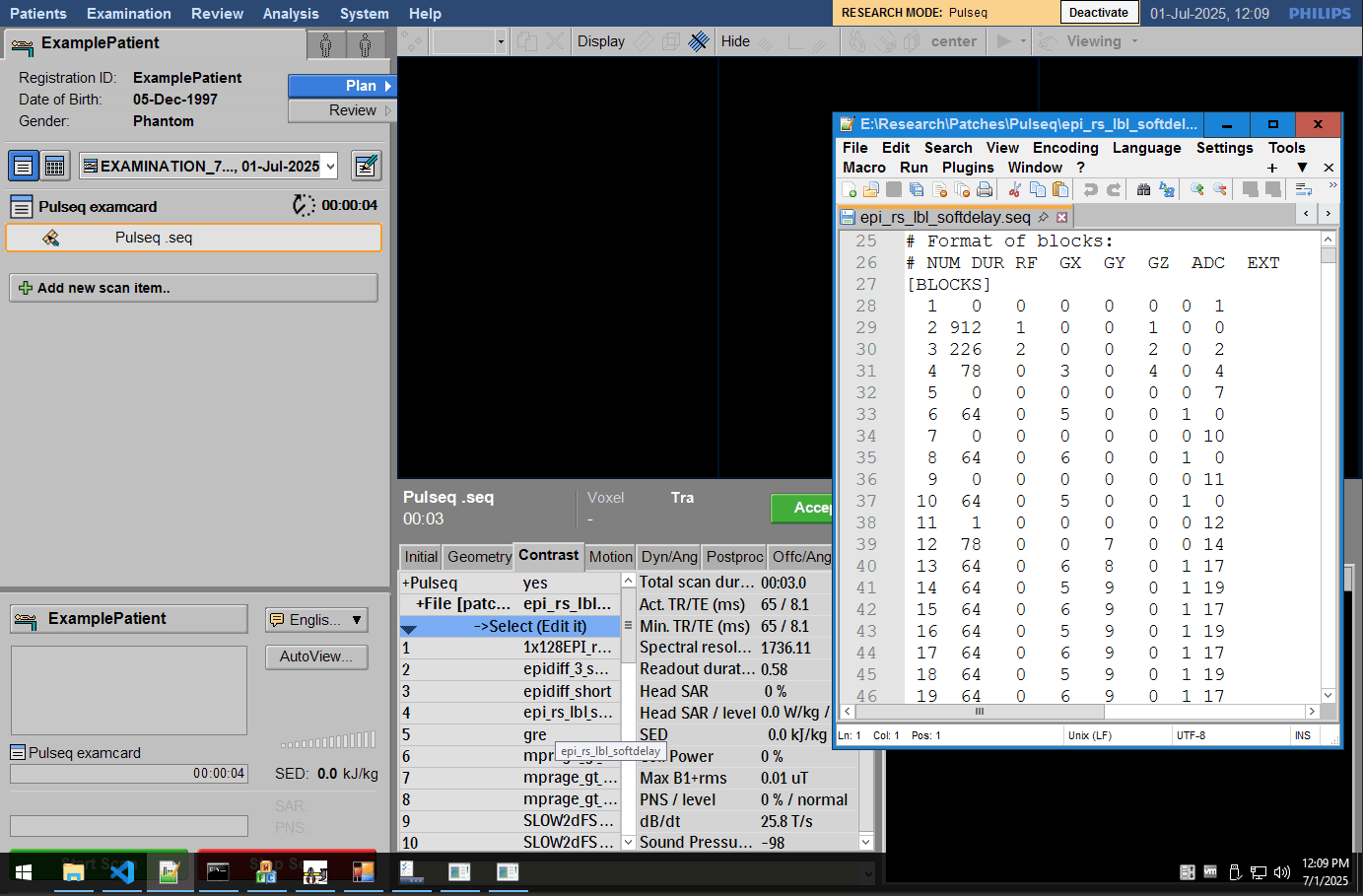
Step 3: Run Your Sequence
Double click on the .ExamCard file provided with the patch source code, to load it into the scanner's database
Next Steps
Now that you're set up, explore what else is possible:
Discover hybrid mode, pTx support, and more.
See FeaturesBrowse complex sequences like EPI and Spiral.
View ExamplesGet help, contribute, and connect with other users.
Get in touch
How to Cite
If you use our Pulseq Interpreter in your research, please cite our work:
BibTeX Citation
@article{Roos2025,
title = {pTx-Pulseq in hybrid sequences: Accessible \& advanced hybrid open-source MRI sequences on Philips scanners},
author = {Thomas H. M. Roos and Edwin Versteeg and Mark Gosselink and Hans Hoogduin and
Kyung Min Nam and Nicolas Boulant and Vincent Gras and Franck Mauconduit and
Dennis W. J. Klomp and Jeroen C. W. Siero and Jannie P. Wijnen},
journal = {Magnetic Resonance in Medicine},
year = {2025},
pages = {1--17},
doi = {10.1002/mrm.30601}
}
APA Style
Roos, T., Versteeg, E., Gosselink, M., Hoogduin, H., Nam, K., Boulant, N., Gras, V., Mauconduit, F., Klomp, D., Siero, J. C. W., & Wijnen, J. (2025).
pTx-Pulseq in hybrid sequences: Accessible & advanced hybrid open-source MRI sequences on Philips scanners.
Magnetic Resonance in Medicine, 1, 1–17.
doi.org/10.1002/mrm.30601